- 23284
- 19880
Journal Club
Omics Data is the Future of Medicine
Feb 1, 2023
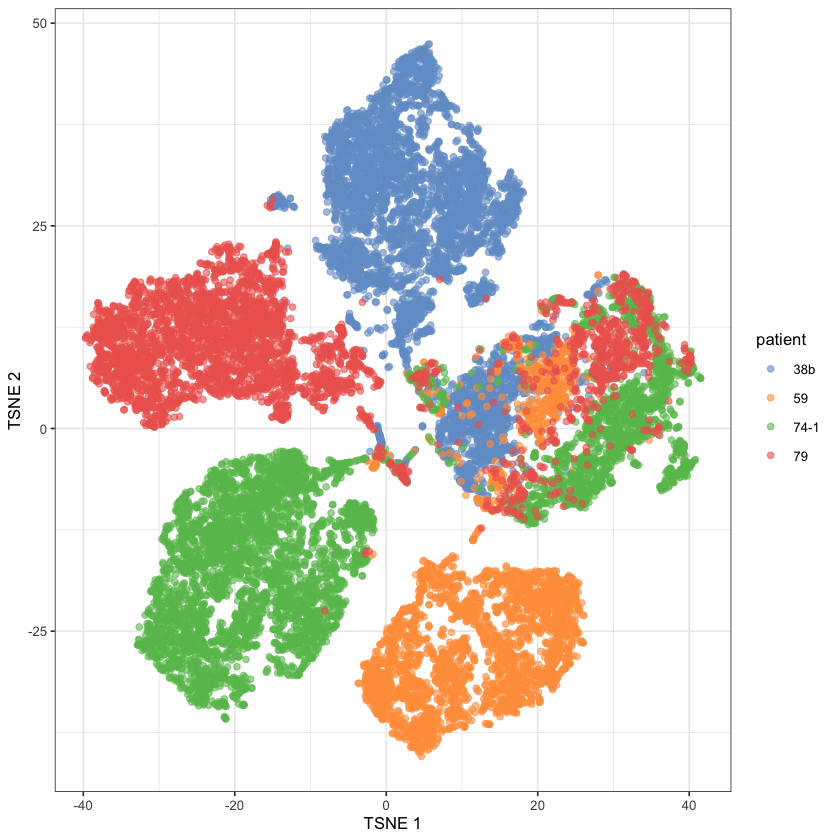
Cancer, from an omics perspective
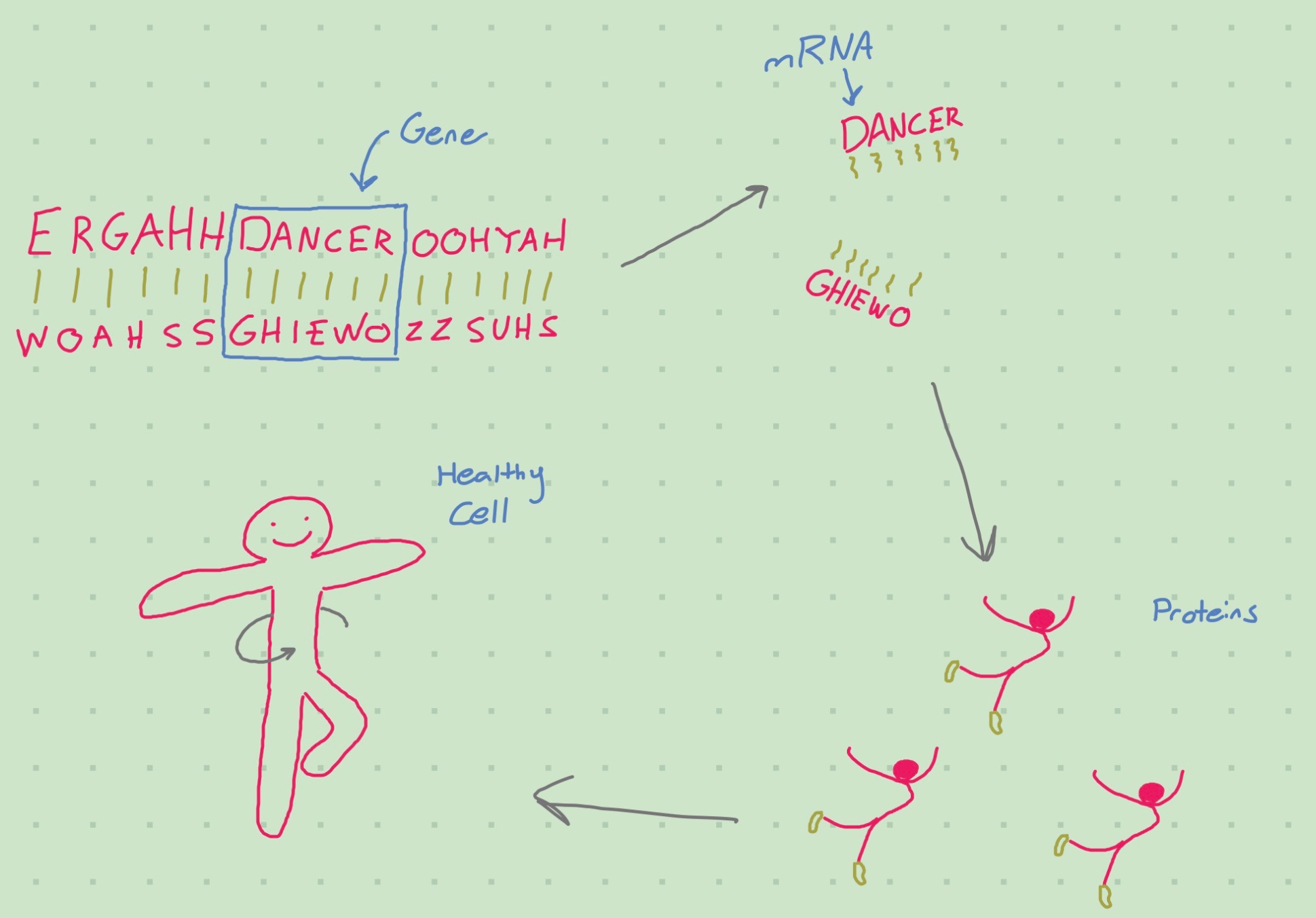
Many ‘omics’: genomics, transcriptomics, proteomics, etc…
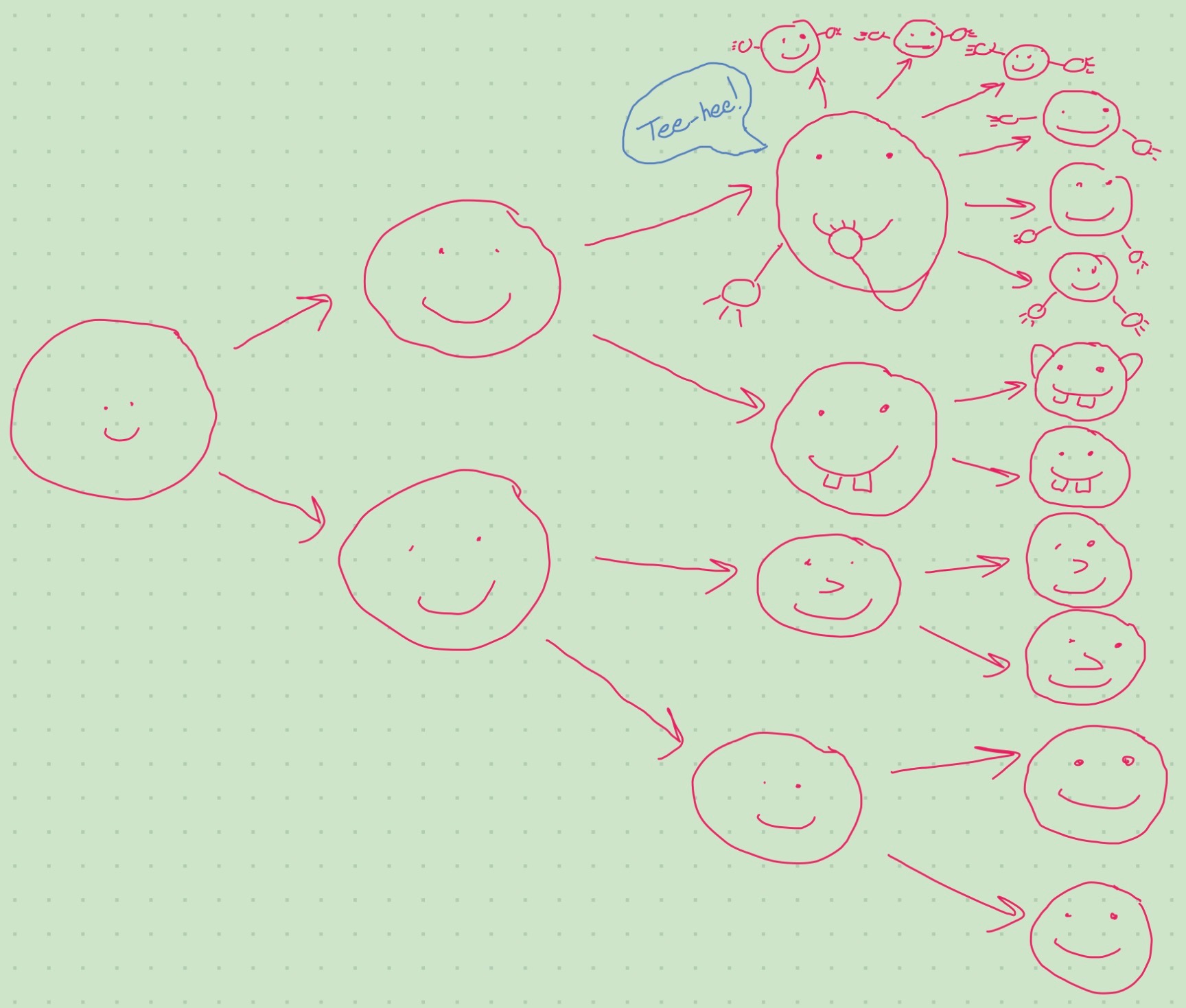
Cells divide and accrue mutations
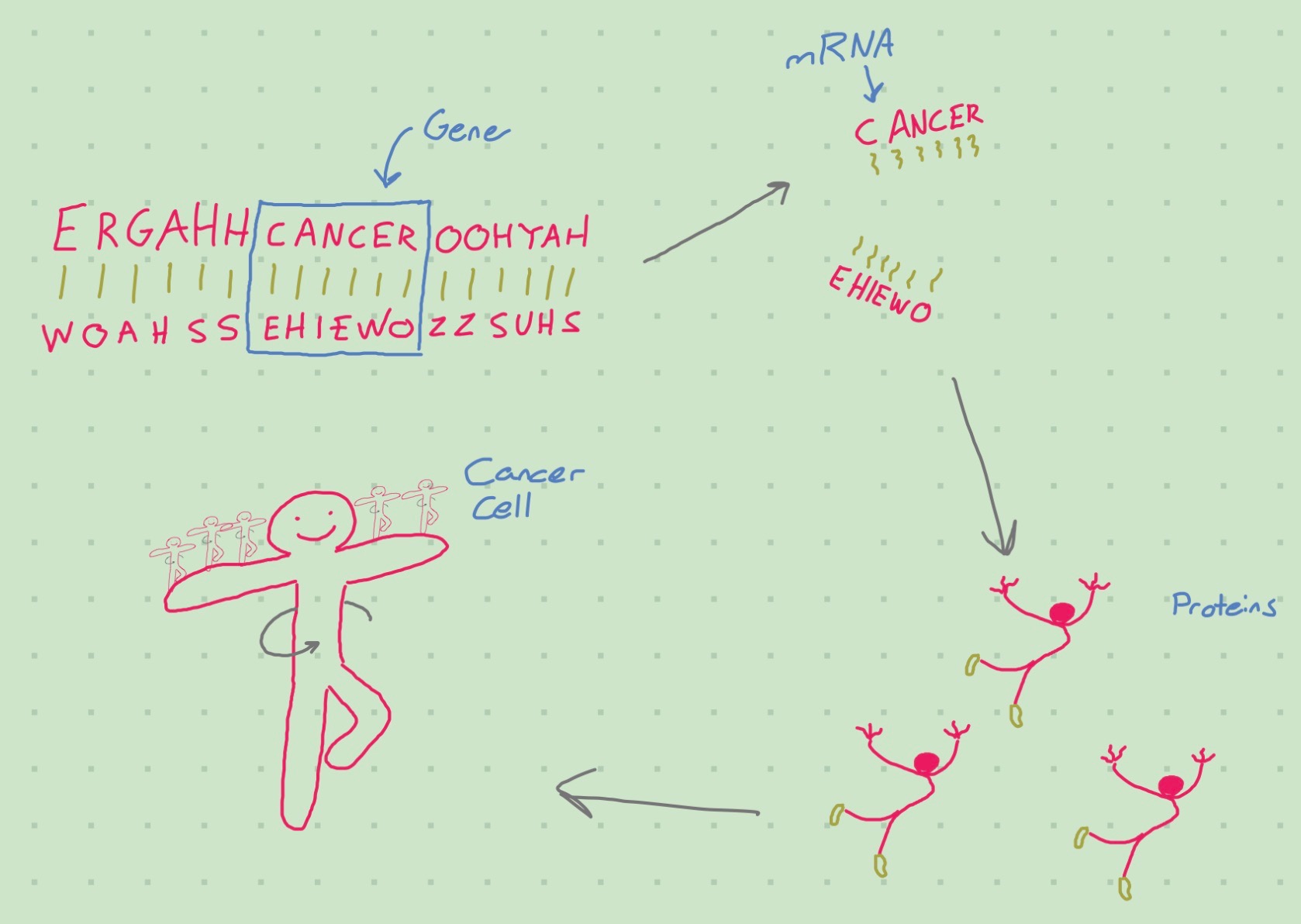
Things can go wrong at the omics level
Why should you care?
- Many diseases are caused by an omics issue
- When so, other data is a proxy1
- heterogenous tumors?: single-cell omics
- Symptoms may be caused by omics issues
- Proteomics enables better design of treatments?
1) Well, environmental factors can be a cause, but limited ability to affect them
Now that you care…
Hands-on Learning Experience™
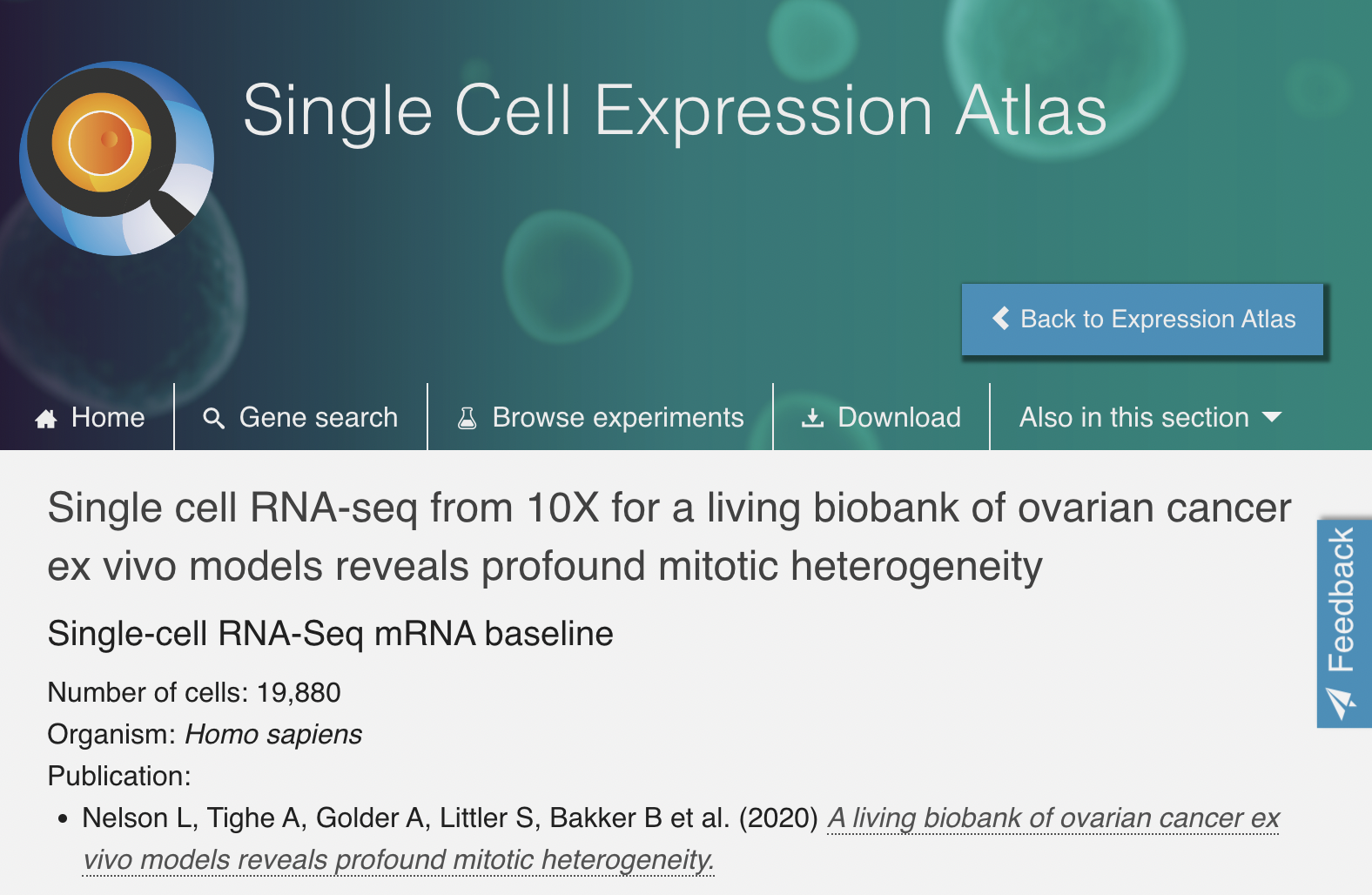
Dataset Details - Where?
To build a living biobank, we established a biopsy pipeline, collecting samples from patients diagnosed with epithelial ovarian cancer treated at the Christie Hospital.
– Nelson et al. (2020)
(Christie Hospital is in Manchester)
Dataset Details - Who?
Between May 2016 and June 2019, we collected 312 samples from patients with chemo-naïve and relapsed disease, either as solid biopsies or as ascites (Fig. 1a)
Ten patients had HGSOC while two had mucinous ovarian carcinoma. Longitudinal biopsies were collected from three patients.
– Nelson et al. (2020)
Dataset Details - How?
The primer contains:
an Illumina TruSeq Read 1 (read 1 sequencing primer)
16 nt 10x Barcode
12 nt unique molecular identifier (UMI)
30 nt poly(dT) sequence
Barcoded, full-length cDNA is amplified via PCR to generate sufficient mass for library construction.
– Nelson et al. (2020)
Load the Data
Load the Data
Ensembl.ID
1 ENSG00000000003
2 ENSG00000000419
3 ENSG00000000457
4 ENSG00000000460
5 ENSG00000000938
6 ENSG00000000971Load the Data
Cell.ID
1 SAMEA6492740-AAACCCACAGTTAGGG
2 SAMEA6492740-AAACCCACATGTGTCA
3 SAMEA6492740-AAACCCAGTCGCATGC
4 SAMEA6492740-AAACCCAGTCTTTCAT
5 SAMEA6492740-AAACCCATCCGTGTCT
6 SAMEA6492740-AAACCCATCCTCTCTTLoad the Metadata
class: SingleCellExperiment
dim: 23284 19880
metadata(0):
assays(1): counts
rownames(23284): ENSG00000000003 ENSG00000000419 ... ENSG00000289701
ENSG00000289716
rowData names(0):
colnames(19880): SAMEA6492740-AAACCCACAGTTAGGG
SAMEA6492740-AAACCCACATGTGTCA ... SAMEA6492743-TTTGTTGGTCCTGGTG
SAMEA6492743-TTTGTTGTCAGATTGC
colData names(1): patient
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):Why So Much Preprocessing?

ELI5: Wet-lab work is hard
class: SingleCellExperiment
dim: 23284 19880
metadata(0):
assays(2): counts logcounts
rownames(23284): ENSG00000000003 ENSG00000000419 ... ENSG00000289701
ENSG00000289716
rowData names(0):
colnames(19880): SAMEA6492740-AAACCCACAGTTAGGG
SAMEA6492740-AAACCCACATGTGTCA ... SAMEA6492743-TTTGTTGGTCCTGGTG
SAMEA6492743-TTTGTTGTCAGATTGC
colData names(2): patient sizeFactor
reducedDimNames(0):
mainExpName: NULL
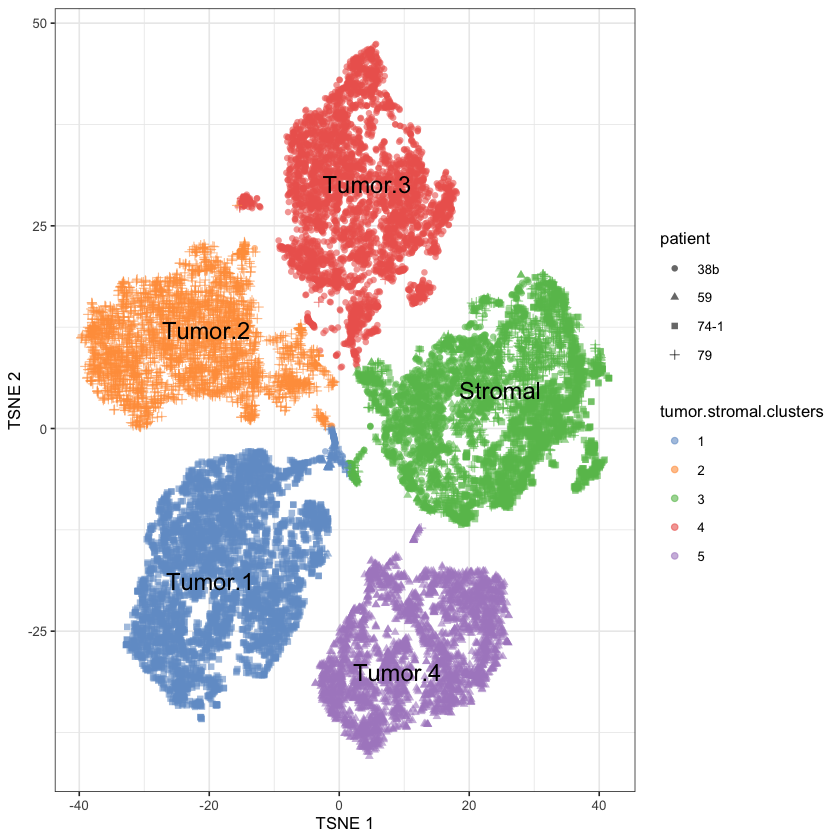
altExpNames(0):Identifying stromals
# We'll assign clusters by eye since it's obvious
ovarian.sce$tumor.or.stromal <- "Stromal"
ovarian.sce$tumor.or.stromal[ovarian.sce$tumor.stromal.clusters==1] <- "Tumor.1"
ovarian.sce$tumor.or.stromal[ovarian.sce$tumor.stromal.clusters==2] <- "Tumor.2"
ovarian.sce$tumor.or.stromal[ovarian.sce$tumor.stromal.clusters==4] <- "Tumor.3"
ovarian.sce$tumor.or.stromal[ovarian.sce$tumor.stromal.clusters==5] <- "Tumor.4"[1] "ENSG00000106366"plotReducedDim(
ovarian.sce,
"TSNE",
colour_by=diff.exp.gene,
shape_by="patient",
text_by="tumor.or.stromal",
text_colour="cyan"
) +
ggtitle("Ovarian Cancer Cells by SERPINE1 (ENSG00000106366) Expression") +
theme(
panel.background = element_rect(fill = "darkblue"),
plot.background = element_rect(fill = "cyan")
)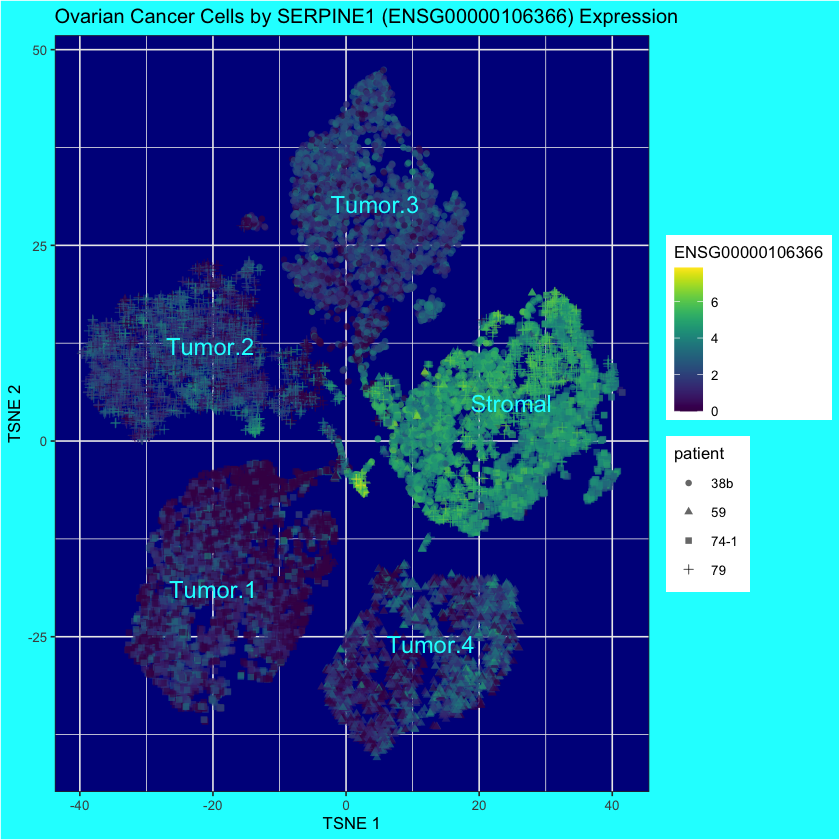
SERPINE1
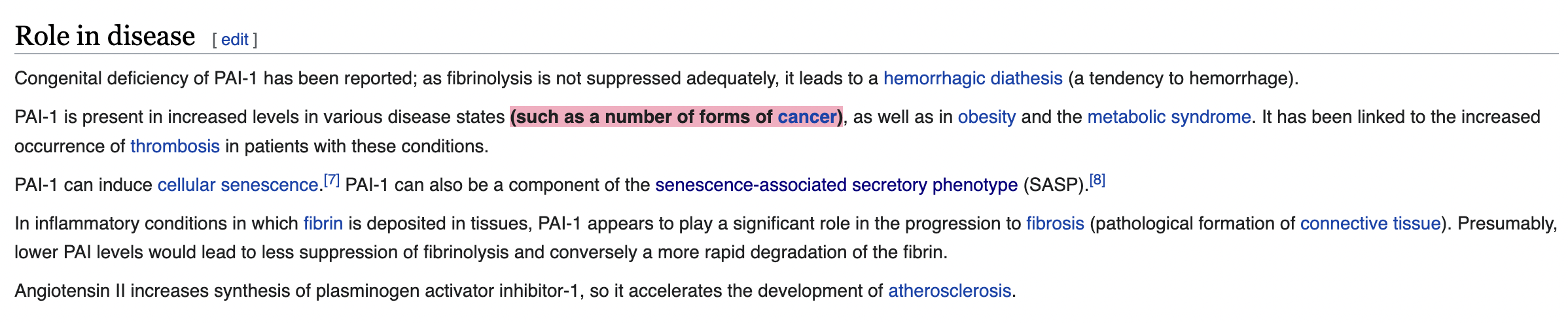
PAI-1, the protein encoded by SERPINE1, is related to cancer!
- Form hypotheses to guide future experiments:
- Can SERPINE1 be a diagnostic factor?
- Is it a cause or symptom of cancer?
- Is it important for tumor health?
- Does it affect patient wellbeing or outcomes?
- Can drugs be designed to target it?
Try It Yourself!
- Go to the Single Cell Expression Atlas
- Pick an interesting dataset
- Download it
- Read the paper
- Read the Bioconductor scRNA ebooks!!!
- Perhaps the best educational resource I’ve used for anything
- Practical and conceptual
- Recreate their preprocessing steps
- Pick a figure in the paper and recreate it
- We recreated Figure 5a from Nelson et al. (2020).